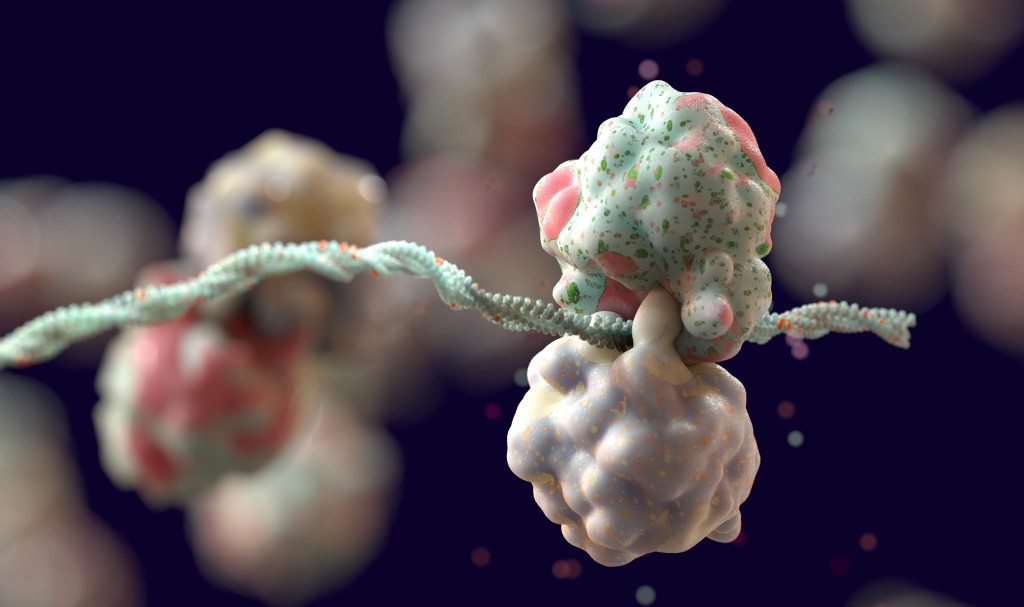
AsianScientist (Feb. 13, 2026) – Ribosomes are the protein factories of our cells, translating messenger RNA (mRNA) into amino acids to synthesise the various proteins that maintain our our bodies.
Liable for a variety of features, proteins catalyse biochemical reactions, act as chemical messengers, and type constructions (together with ribosomes) to make up our our bodies.
Since proteins are so vital, it’s vital that ribosomes perform effectively and precisely. Till now, scientists haven’t understood how cells can detect inefficient ribosomes.
A crew of researchers from the College of Tokyo and Tohoku College have recognized a top quality management mechanism by which cells mark much less environment friendly ribosomes for removing. Printed in Nature Communications, this research discovered that much less environment friendly ribosomes are selectively eliminated when there are higher ribosomes current within the cell.
The researchers engineered yeast cells to include a further, suboptimal ribosome variant that strikes extra slowly than the native ribosomes within the cell. They then used biochemical and genetic analyses to look at the impact of disrupted translation carried out by the suboptimal ribosomes.
Ribosomes work by travelling alongside a string of mRNA as they ‘learn’ it, translating every codon right into a corresponding amino acid. When ribosomes within the yeast cells translated the identical strand of messenger RNA, the slower, suboptimal ribosomes had been overtaken by the native ribosomes, inflicting the 2 to collide
“Ribosome collisions act like a mobile warning sign,” mentioned Assistant Professor Sihan Li, lead writer of the paper. “When ribosomes stumble upon one another, it alerts the cell that one thing is fallacious. The cell then removes the problematic ribosomes to take care of environment friendly protein manufacturing.”
The colliding ribosomes activate a top quality management pathway that triggers ubiquitination. A small protein referred to as ubiquitin attaches to one of many proteins that make up the ribosome, marking it for degradation or high quality management.
“Our research introduces the idea of ribosome competitors, exhibiting that even purposeful however slower ribosomes are selectively degraded when extra succesful ones can be found,” mentioned Dr Li. “It’s fascinating to see how cells apply a precept just like survival of the fittest on the molecular stage.”
The researchers additionally explored the consequences of cisplatin on the ribosome high quality management course of. Cisplatin is a extensively used most cancers drug that interacts with genetic materials, so the scientists hypothesised that this may disrupt ribosome dynamics by linking completely different RNA varieties collectively.
Cisplatin was discovered to extend ribosome collisions, thus activating the ubiquitination pathway and rising the speed of ribosome degradation. This might assist clarify a few of cisplatin’s unintended effects, and improves our understanding of the drug’s toxicity and efficacy.
These outcomes present perception into the standard management course of for ribosomes, and will assist us perceive issues attributable to ribosome malfunctions (ribosomopathies). As seen with cisplatin, this may increasingly additionally open new approaches to enhance the effectiveness of some medication.
“We hope folks discover it attention-grabbing that even on the microscopic stage, competitors drives high quality and resilience,” mentioned Dr. Li. “This discovery connects a basic evolutionary thought to the interior workings of our cells.”
—
Supply: College of Tokyo;Picture:Steven McDowell/Shutterstock
This text might be discovered at: Collision-induced ribosome degradation pushed by ribosome competitors and translational perturbations
Disclaimer: This text doesn’t essentially replicate the views of AsianScientist or its employees.